6.03 DNA Replication
Overview of DNA Replication
- Process: DNA replication is the process of copying DNA, ensuring that each daughter cell receives an identical set of genetic material.
- Timing: Occurs during the S phase of the cell cycle.
- Enzyme Control: Controlled by enzymes, primarily DNA polymerase and DNA ligase.

Steps in DNA Replication
1. Initiation
- Each original strand serves as a template for a new strand.
- Primase: Synthesizes short RNA primers on both strands, providing a starting point for DNA polymerase.
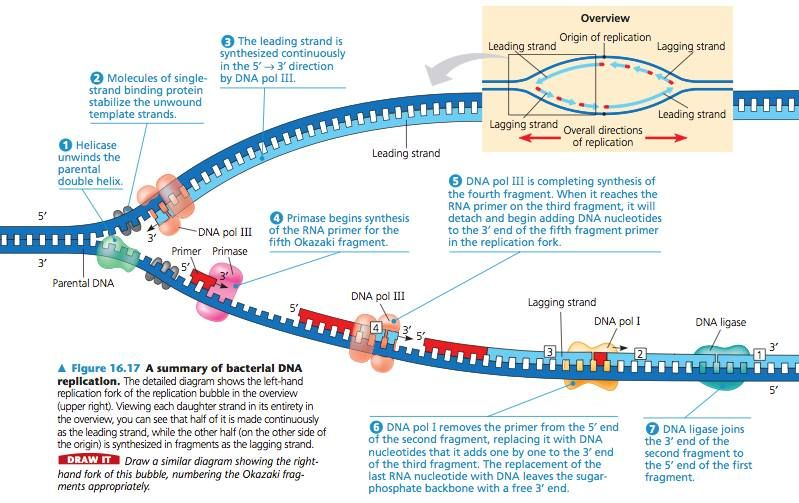
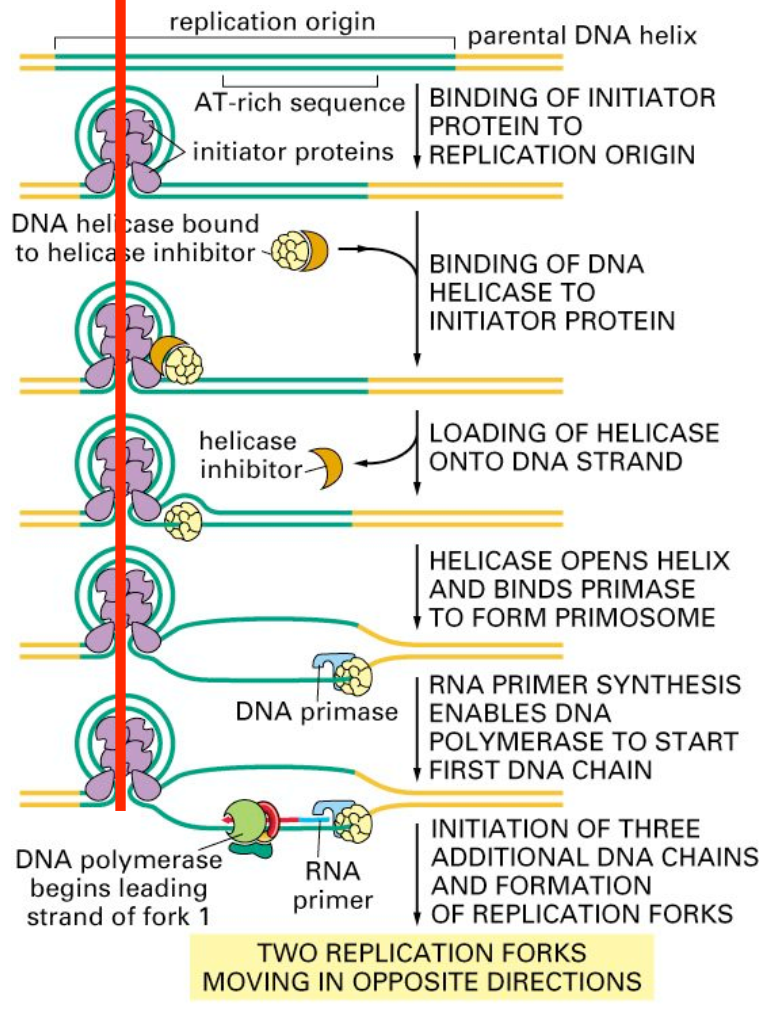
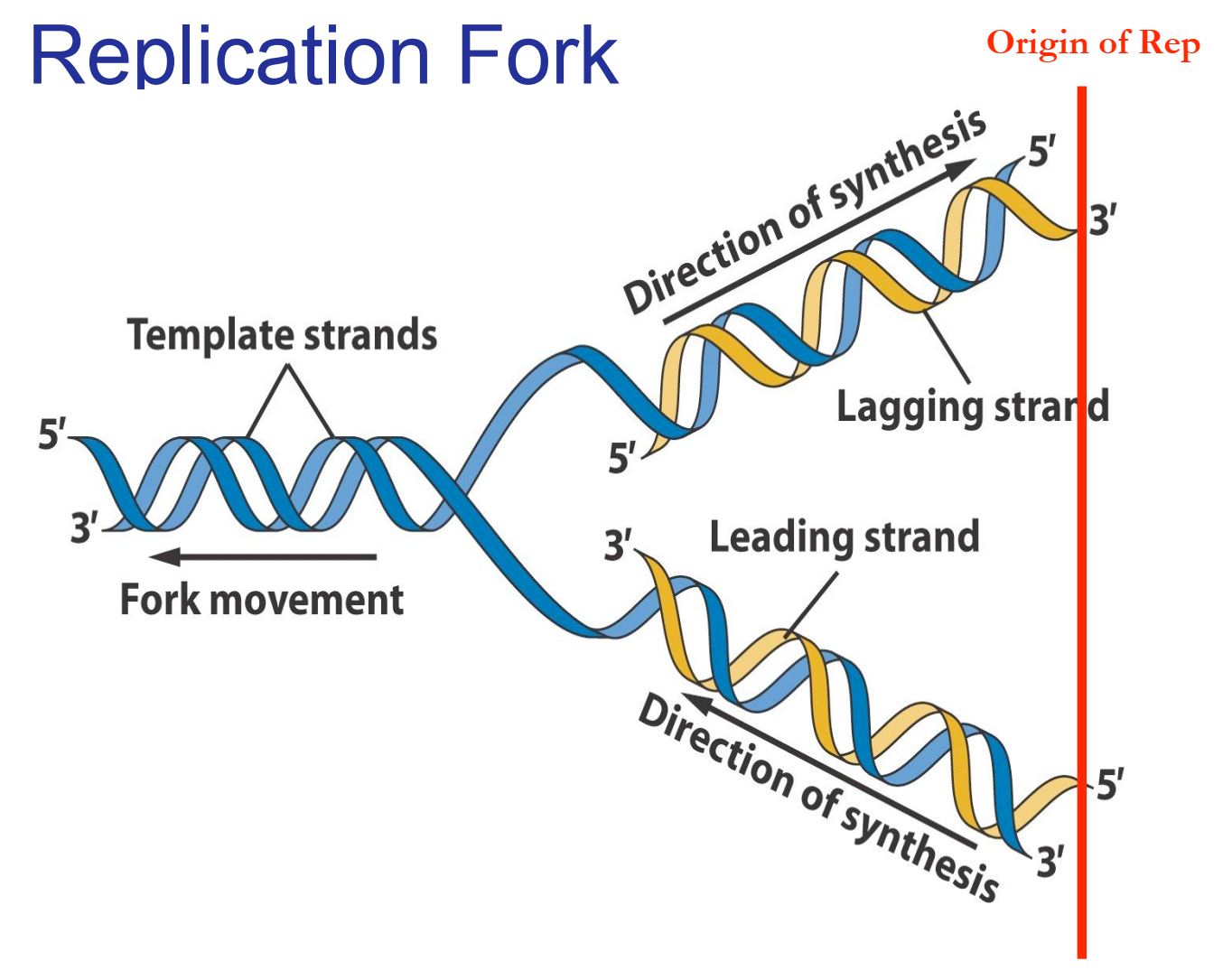
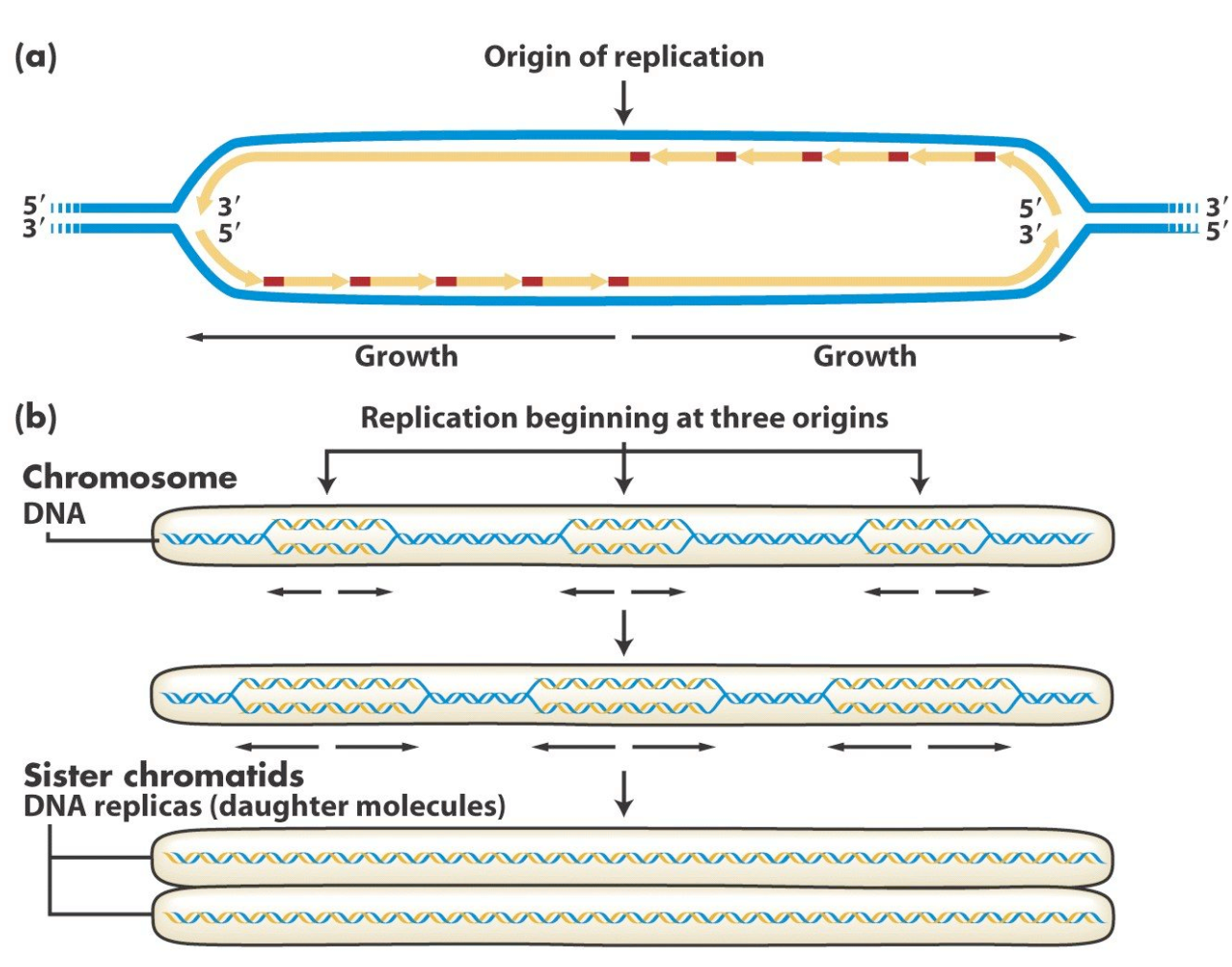
- Origin of Replication: Replication begins at specific sequences called origins of replication.
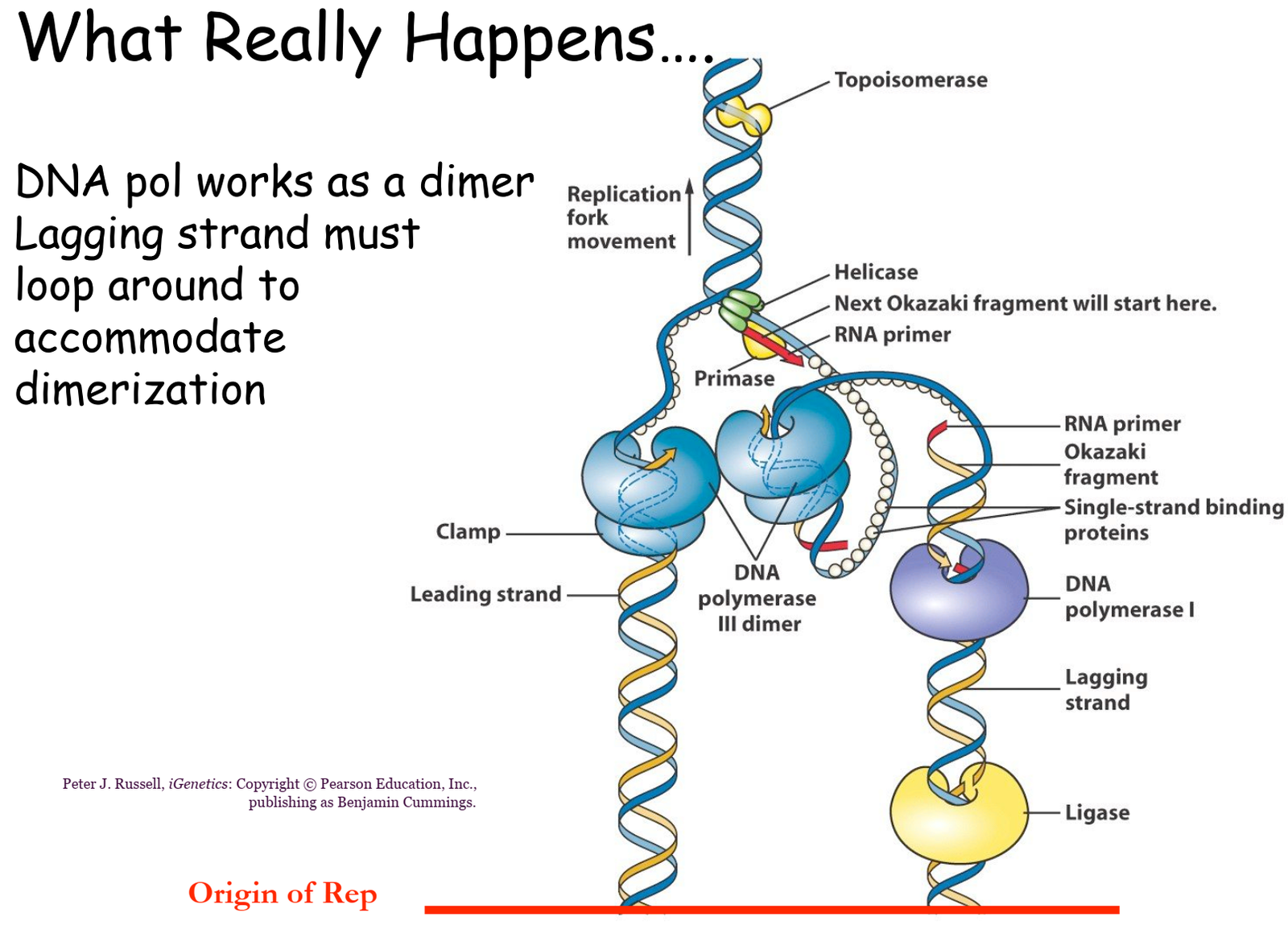
- Helicase Activation: Its primary function is to unwind the double-stranded DNA by breaking the hydrogen bonds between complementary base pairs.
- Single-Strand Binding Proteins (SSBs): These stabilize the separated strands, preventing them from reannealing.
- Topoisomerase: Relieves supercoiling (overwinding) ahead of the replication fork by cutting and rejoining DNA strands.
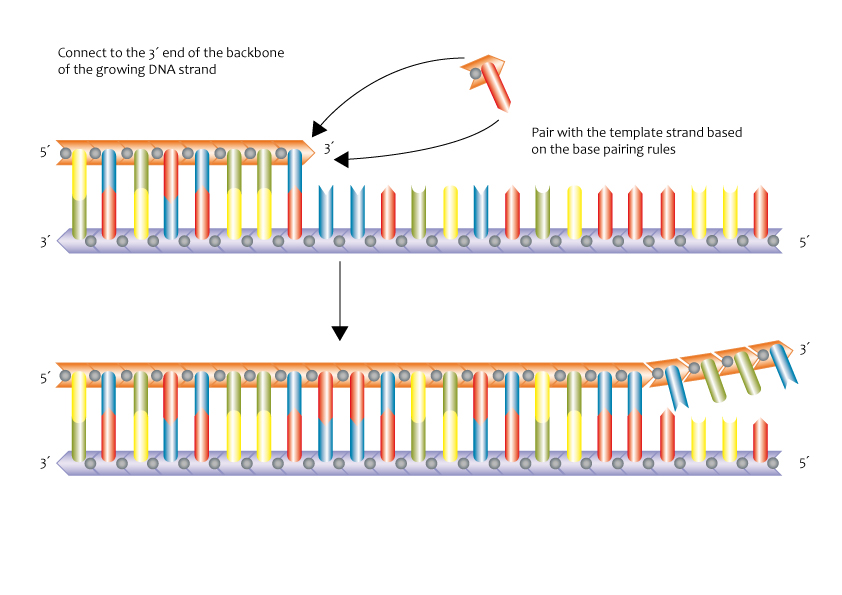
2. Elongation
- Nucleotide Addition: Free nucleotides pair with complementary bases (A-T, G-C), and DNA polymerase catalyzes the formation of phosphodiester bonds.
- DNA Polymerase binds to each of the separated strands.
- DNA Polymerase – Leading Strand Synthesis:
- DNA polymerase adds complementary nucleotides continuously in the 5’ to 3’ direction (towards the replication fork).
- DNA Polymerase – Lagging Strand Synthesis:
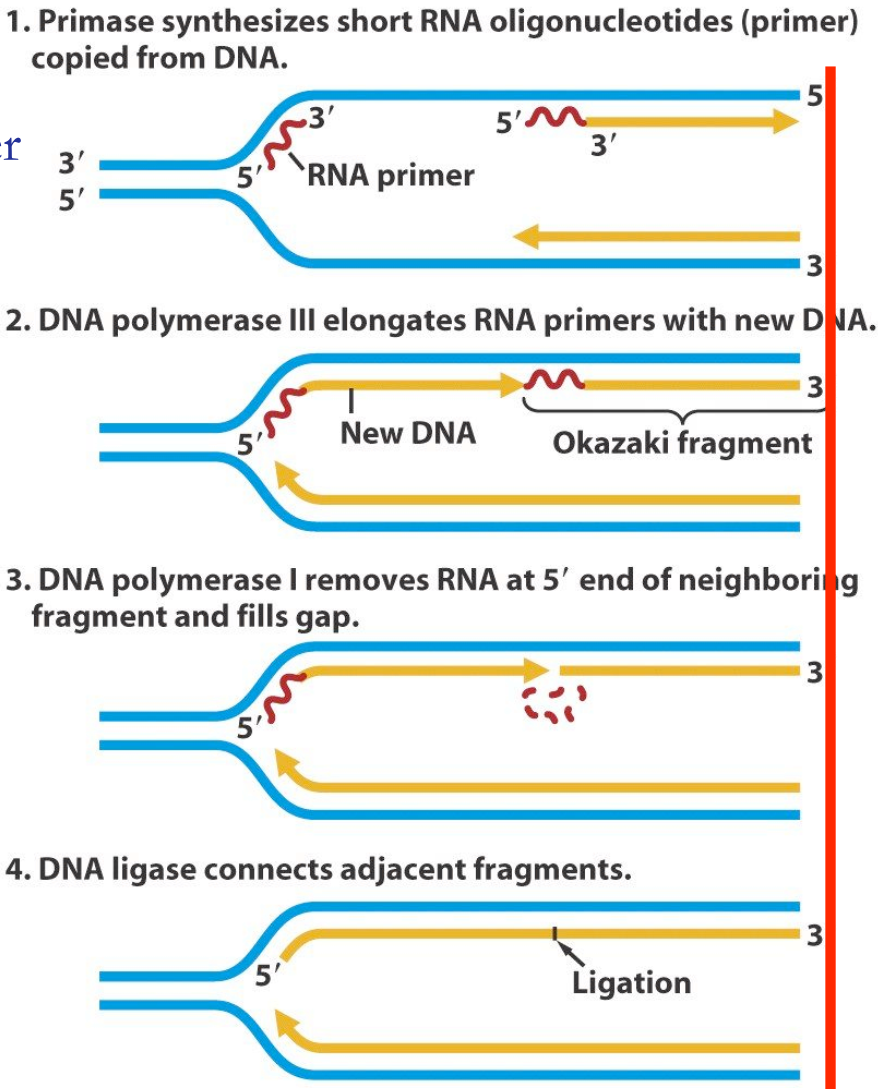
- DNA polymerase synthesizes DNA discontinuously as short fragments called Okazaki fragments in the 5’ to 3’ direction (away from the replication fork).
- Each fragment starts with an RNA primer laid down by primase.
- DNA Polymerase – Leading Strand Synthesis:
3. Termination
- Primer Removal:
- RNA primers are removed by DNA polymerase I.
- Gaps left by the removal of primers are filled with DNA by DNA polymerase.
- Okazaki Fragment Ligation:
- DNA ligase joins the Okazaki fragments on the lagging strand.
- Forms covalent phosphodiester bonds to complete the sugar-phosphate backbone, linking Okazaki fragments into a continuous strand.
- Replication Fork Completion:
- Replication forks meet at specific termination sites, or when the entire DNA molecule has been replicated.

4. Proofreading and Error Correction
- DNA Polymerase Proofreading:
- DNA polymerase checks for mismatched bases during replication and corrects errors using its 3’ to 5’ exonuclease activity.
- Post-Replication Repair:
- Additional repair mechanisms correct any remaining errors or damage after replication.
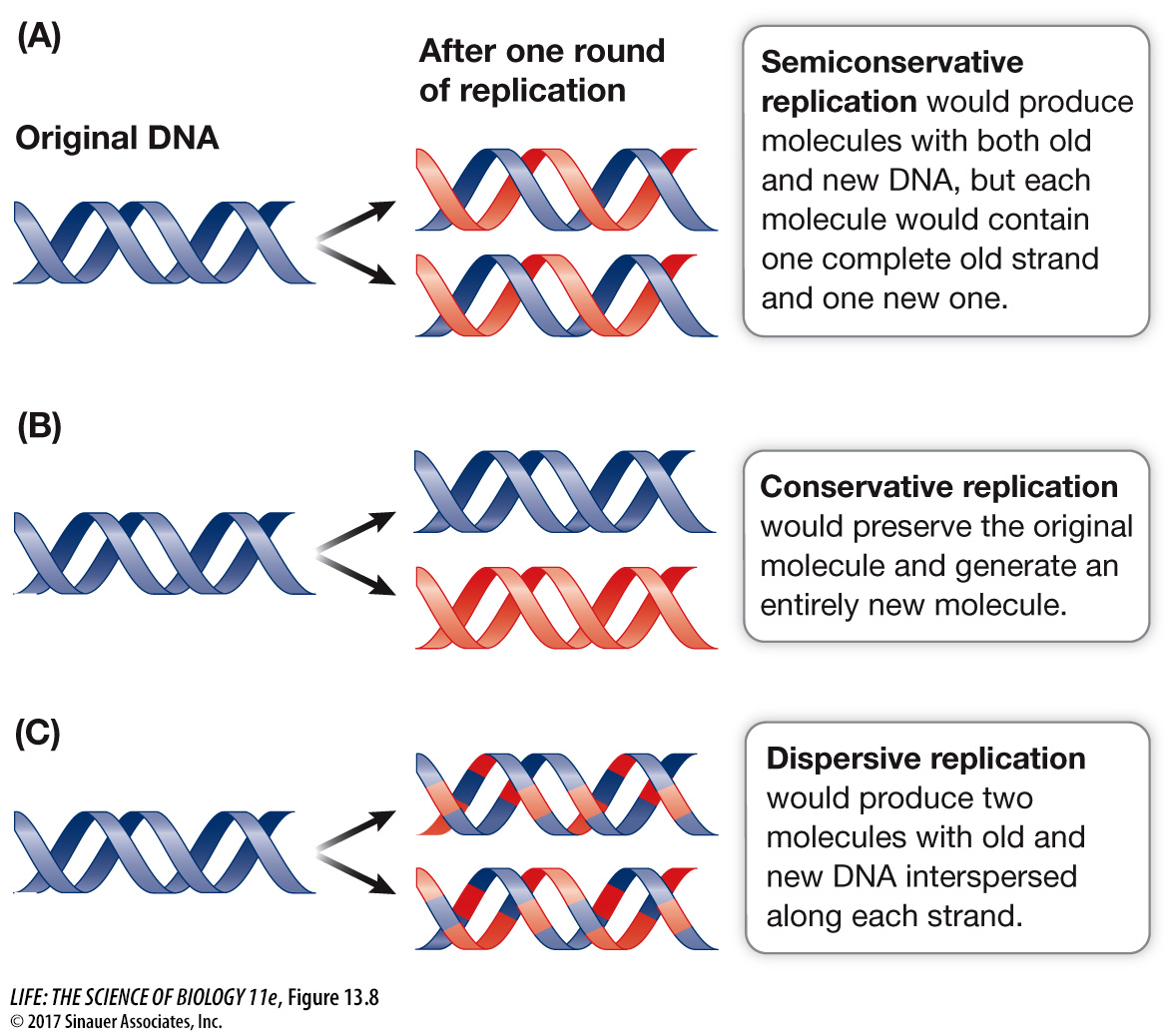
Semi-Conservative Replication
- Definition: Each new DNA molecule consists of one original (parent) strand and one newly synthesized strand.
- The original strands are conserved in each of the new DNA molecules, hence “semi-conservative.”
- Alternative Hypothesis (Conservative Replication): If DNA replication were conservative, the entire parent DNA molecule would remain intact, and the new DNA molecule would have two new strands.
- Alternative Hypothesis (Dispersive Replication) : If DNA replication were dispersive, the parent DNA molecule would be fragmented, and each daughter DNA molecule would be a mix of old (parental) and new DNA. Both strands of the daughter DNA would contain alternating segments of old and newly synthesized DNA throughout their length.
Key Molecules in DNA Replication
1. DNA Polymerase:
- Function: Adds complementary nucleotides to the template strands in the 5′ to 3′ direction.
- Process:
- Leading strand: Synthesized continuously.
- Lagging strand: Synthesized in short Okazaki fragments.
2. DNA Ligase:
- Function: Joins nucleotides together to form the continuous DNA backbone.
- Process: Links Okazaki fragments on the lagging strand and secures phosphodiester bonds between adjacent nucleotides.
3. Other Necessary Molecules:
- Nucleotides: The building blocks for new DNA strands.
- Helicase: An enzyme that separates DNA strands (initiates unwinding).
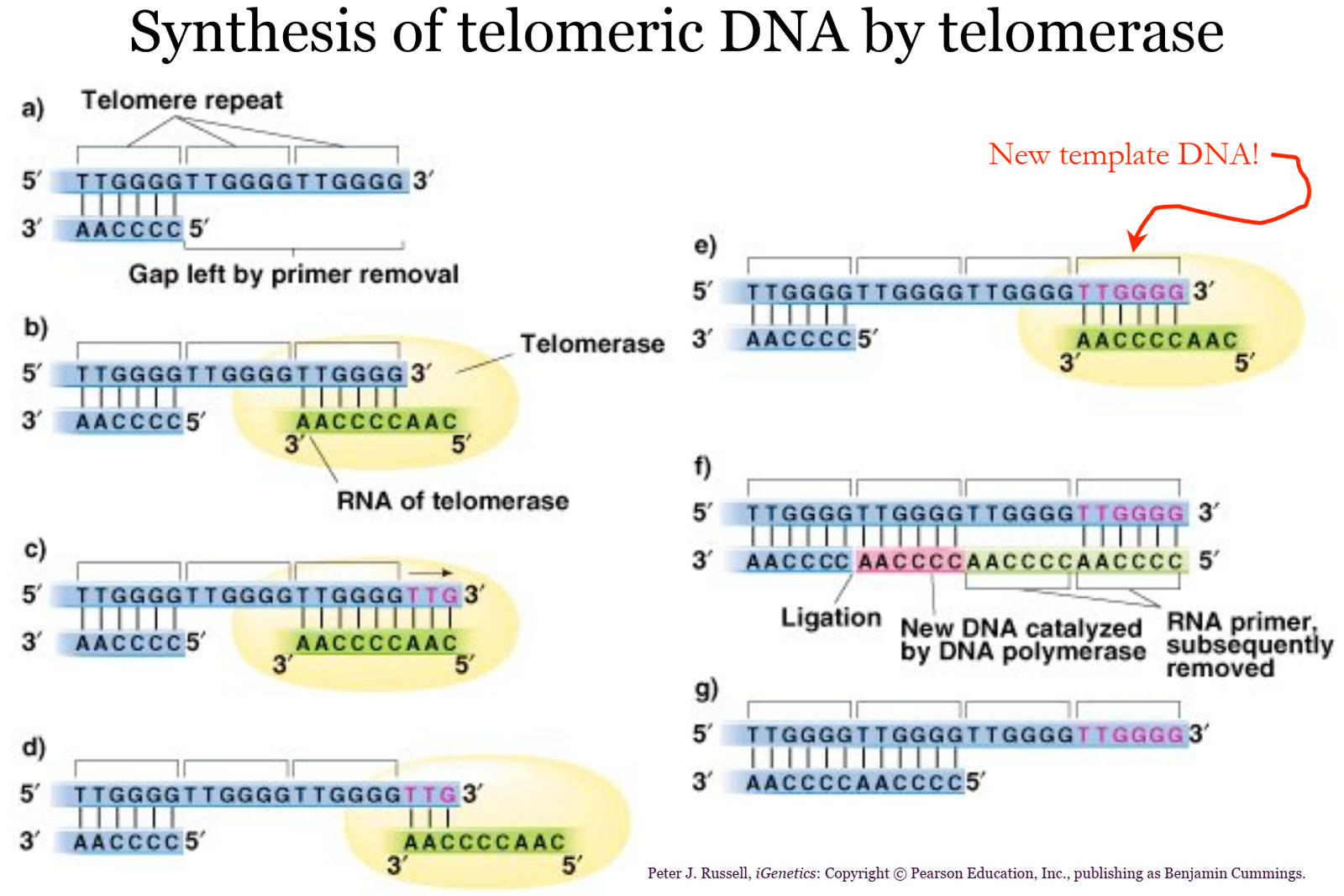
DNA Polymerase and Telomere Replication
- DNA Polymerase Functionality:
- DNA polymerase synthesizes DNA in a 5’ to 3’ direction by adding nucleotides to the 3’-OH end of the growing strand.
- It requires a primer to initiate replication but cannot begin synthesis on its own.
- Problem at the Ends (Lagging Strand):
- At the ends of linear chromosomes, DNA polymerase faces difficulty replicating the telomeres (repetitive DNA sequences at chromosome ends).
- On the lagging strand, RNA primers are required for Okazaki fragments. Once the primer at the very end is removed, there is no upstream 3’-OH group for DNA polymerase to extend.
- This leads to the end-replication problem, where the lagging strand cannot be fully replicated, resulting in gradual telomere shortening during successive cell divisions.
- Role of Telomerase:
- Telomerase is a specialized enzyme that resolves the end-replication problem by adding repetitive sequences to the ends of chromosomes.
- It contains an RNA template that it uses to extend the 3’ end of the parental strand, providing a template for DNA polymerase to complete lagging strand synthesis.
- Telomerase is active in germ cells, stem cells, and certain cancer cells but is typically inactive in most somatic cells.
- Consequences of Telomere Shortening:
- In the absence of telomerase, telomeres progressively shorten with each round of replication.
- Critically short telomeres signal the cell to enter senescence (growth arrest) or undergo apoptosis (programmed cell death), limiting the lifespan of the cell.
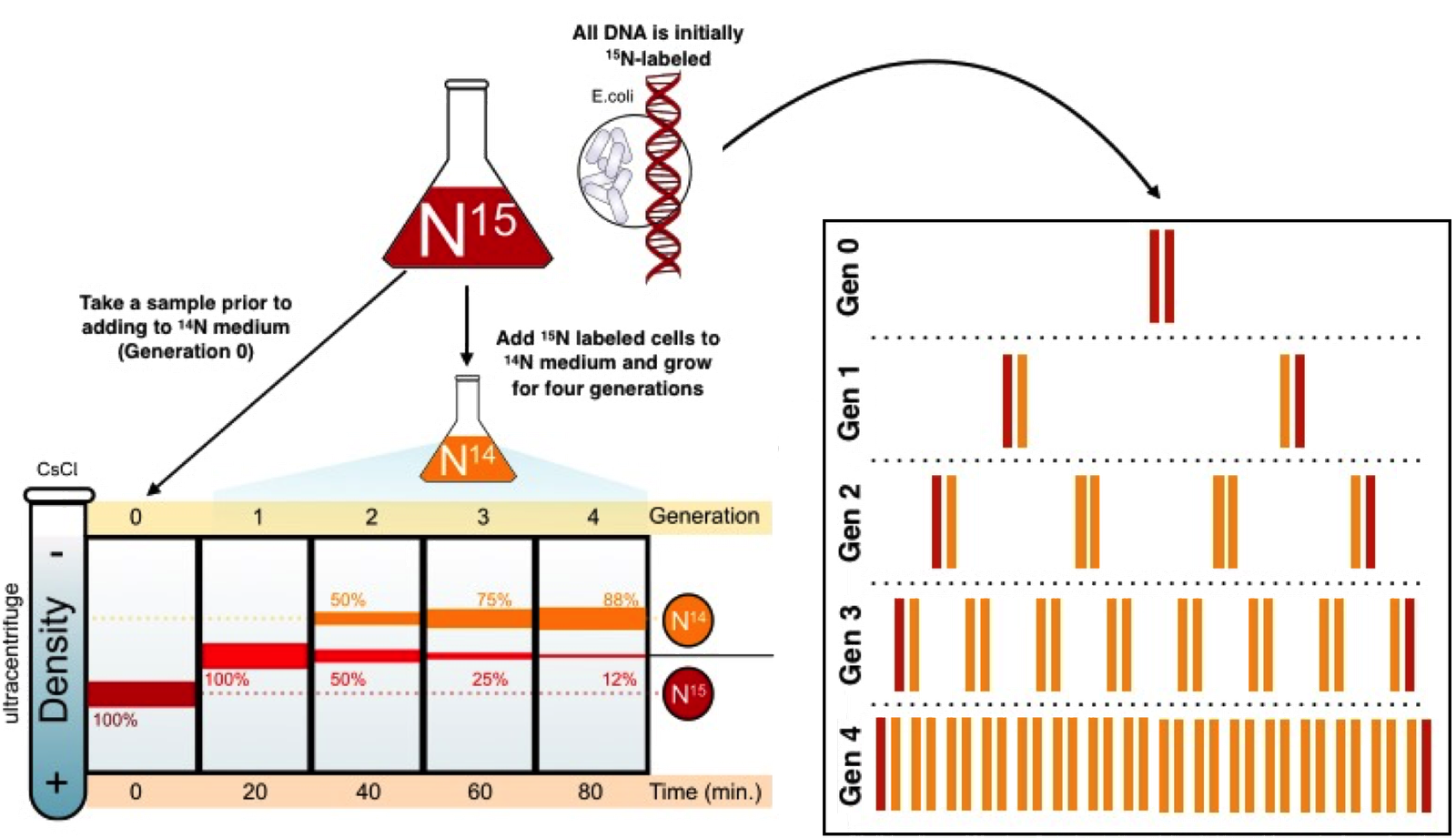
Experiment to Prove Semi-Conservative Replication (Meselson & Stahl, 1958)
Objective:
- Determine whether DNA replication is semi-conservative, conservative, or dispersive.
Experimental Design:
- Bacterial System Used:
- E. coli grown in a medium containing heavy nitrogen isotope (15N15N) to label DNA.
- Procedure:
- Step 1: Incorporation of 15N15N:
- E. coli grown in 15N15N medium → DNA became fully labeled with heavy nitrogen.
- Step 2: Switch to 14N14N:
- Cells transferred to a medium with light nitrogen isotope (14N14N).
- Step 3: Density Gradient Centrifugation:
- DNA extracted after successive replication cycles.
- DNA density analyzed using cesium chloride (CsCl) gradient centrifugation.
- Step 1: Incorporation of 15N15N:
Results:
- Generation 0 (before replication):
- DNA entirely heavy (15N15N) → formed a single, dense band.
- Generation 1 (after one replication cycle in 14N14N):
- DNA formed intermediate density band (hybrid of 15N15N and 14N14N).
- Ruled out conservative replication (would produce one heavy and one light band).
- Generation 2 (after two replication cycles in 14N14N):
- DNA formed two bands:
- One light band (14N14N).
- One intermediate band (15N/14N 15N/14N).
- Ruled out dispersive replication (would only produce intermediate bands).
- DNA formed two bands:
Conclusion:
- DNA replication is semi-conservative:
- Each daughter molecule contains one original (parental) strand and one newly synthesized strand.
Additional Explanation:
Labeling DNA:
- The bacteria were first grown in a medium containing ¹⁵N (heavy nitrogen), so their DNA became fully labeled with the heavier nitrogen isotope.
- Then, the bacteria were transferred to a medium with ¹⁴N (light nitrogen), and they were allowed to replicate their DNA.
Harvesting DNA:
- After one or more rounds of replication, DNA samples were extracted from the bacteria.
Density Gradient Centrifugation:
- The DNA samples were then subjected to density gradient centrifugation, a technique that uses a solution of cesium chloride (CsCl) to create a gradient in which DNA molecules can separate based on their density.
- DNA molecules containing ¹⁵N (heavy nitrogen) would form a band lower in the gradient, while DNA molecules containing ¹⁴N (light nitrogen) would form a band higher in the gradient.
Detection:
- The researchers used radioactive isotope detectors to detect the different densities of the DNA.
- The ¹⁵N-labeled DNA was more dense than the ¹⁴N-labeled DNA, so when the DNA was centrifuged, the bands could be detected at different positions in the gradient.
Results:
- After one round of replication in the presence of ¹⁴N, the DNA showed a single intermediate band, which was consistent with semi-conservative replication (where one strand is from the parent DNA and the other is newly synthesized using ¹⁴N).
- After two rounds of replication, the DNA showed two bands: one band of intermediate density (from hybrid molecules containing one strand of ¹⁵N and one strand of ¹⁴N) and one band of light DNA (¹⁴N-only DNA).
Conclusion:
- The results confirmed that DNA replication is semi-conservative, meaning that each new DNA molecule consists of one strand from the original (parental) DNA and one newly synthesized strand.
Practise Questions
Question 1
Explain the role of DNA polymerase and DNA ligase in DNA replication. (6 marks)
Mark Scheme:
- DNA polymerase synthesizes DNA by adding complementary nucleotides to the template strand in the 5′ to 3′ direction. (1 mark)
- It catalyzes the formation of phosphodiester bonds between adjacent nucleotides. (1 mark)
- On the leading strand, DNA polymerase synthesizes DNA continuously. (1 mark)
- On the lagging strand, it synthesizes short fragments called Okazaki fragments, requiring multiple primers. (1 mark)
- DNA ligase joins the Okazaki fragments on the lagging strand, forming covalent phosphodiester bonds. (1 mark)
- This ensures the formation of a continuous sugar-phosphate backbone in the lagging strand. (1 mark)
Question 2
Describe the process of semi-conservative DNA replication. (6 marks)
Mark Scheme:
- DNA replication begins with the unwinding of the double helix by helicase, which breaks hydrogen bonds between complementary base pairs. (1 mark)
- Each strand serves as a template for the synthesis of a new complementary strand. (1 mark)
- Primase synthesizes RNA primers, which provide a starting point for DNA polymerase. (1 mark)
- DNA polymerase adds nucleotides complementary to the template strand (A-T, G-C) in the 5′ to 3′ direction. (1 mark)
- On the leading strand, DNA is synthesized continuously, while on the lagging strand, it is synthesized discontinuously as Okazaki fragments. (1 mark)
- The result is two DNA molecules, each consisting of one original strand and one newly synthesized strand, demonstrating semi-conservative replication. (1 mark)
Question 3
What are the roles of helicase, single-strand binding proteins (SSBs), and topoisomerase in DNA replication? (6 marks)
Mark Scheme:
- Helicase unwinds the DNA double helix by breaking hydrogen bonds between base pairs. (1 mark)
- This creates two single strands, forming the replication fork. (1 mark)
- SSBs stabilize the separated strands, preventing them from reannealing or forming secondary structures. (1 mark)
- Topoisomerase relieves supercoiling ahead of the replication fork by cutting and rejoining DNA strands. (1 mark)
- This prevents tension buildup that could hinder replication. (1 mark)
- These enzymes ensure the DNA is properly unwound and stabilized for replication to proceed efficiently. (1 mark)
Question 4
Describe how Okazaki fragments are synthesized and joined during DNA replication. (5 marks)
Mark Scheme:
- On the lagging strand, DNA is synthesized discontinuously in short fragments called Okazaki fragments. (1 mark)
- Primase synthesizes an RNA primer for each fragment, providing a starting point for DNA polymerase. (1 mark)
- DNA polymerase extends the primer by adding nucleotides in the 5′ to 3′ direction, away from the replication fork. (1 mark)
- Once the RNA primers are removed by DNA polymerase I, the gaps are filled with DNA. (1 mark)
- DNA ligase joins the fragments, forming phosphodiester bonds to create a continuous DNA strand. (1 mark)
Question 5
What evidence did Meselson and Stahl provide to support the semi-conservative model of DNA replication? (6 marks)
Mark Scheme:
- Objective: Determine whether DNA replication is semi-conservative, conservative, or dispersive. (1 mark)
- Bacteria were grown in a medium containing 15N (heavy nitrogen), which labeled their DNA with a heavier isotope. (1 mark)
- After one round of replication in 14N (light nitrogen), the DNA formed a single intermediate-density band, ruling out conservative replication. (1 mark)
- After two rounds of replication in 14N, two bands appeared: one light band and one intermediate band. (1 mark)
- This ruled out dispersive replication, which would produce only intermediate bands. (1 mark)
- The results confirmed the semi-conservative model, where each daughter DNA molecule consists of one original strand and one newly synthesized strand. (1 mark)
Question 6
Explain the role of telomerase in DNA replication and its importance in certain cell types. (6 marks)
Mark Scheme:
- Telomerase is an enzyme that extends the telomeres, repetitive DNA sequences at chromosome ends. (1 mark)
- It uses its RNA template to add nucleotides to the 3′ end of the parental DNA strand. (1 mark)
- This provides a template for DNA polymerase to complete the lagging strand synthesis. (1 mark)
- Telomerase is active in germ cells, stem cells, and cancer cells, enabling these cells to divide indefinitely. (1 mark)
- In most somatic cells, telomerase is inactive, leading to gradual telomere shortening and limiting cell lifespan. (1 mark)
- Telomerase prevents telomere shortening, which would otherwise trigger senescence or apoptosis. (1 mark)
Question 7
What is the significance of proofreading during DNA replication, and how does DNA polymerase correct errors? (5 marks)
Mark Scheme:
- DNA polymerase has proofreading ability to ensure high fidelity during DNA replication. (1 mark)
- It detects mismatched bases by recognizing structural irregularities in the DNA. (1 mark)
- The 3′ to 5′ exonuclease activity of DNA polymerase removes the incorrect nucleotide. (1 mark)
- DNA polymerase then adds the correct complementary nucleotide. (1 mark)
- This proofreading reduces replication errors, maintaining the integrity of the genetic information. (1 mark)
Question 8
Compare the challenges of replicating the leading strand versus the lagging strand. (5 marks)
Mark Scheme:
- The leading strand is synthesized continuously in the 5′ to 3′ direction toward the replication fork. (1 mark)
- This makes replication straightforward, with DNA polymerase moving in the same direction as helicase. (1 mark)
- The lagging strand is synthesized discontinuously in the 5′ to 3′ direction away from the replication fork. (1 mark)
- This requires the synthesis of multiple Okazaki fragments, each starting with an RNA primer. (1 mark)
- Joining these fragments with DNA ligase adds complexity to lagging strand replication. (1 mark)
Question 9
Why does telomere shortening occur in somatic cells, and what are its consequences? (5 marks)
Mark Scheme:
- Telomere shortening occurs because DNA polymerase cannot fully replicate the ends of linear chromosomes. (1 mark)
- The removal of RNA primers at the chromosome ends leaves a gap that cannot be filled. (1 mark)
- With each replication cycle, telomeres become progressively shorter. (1 mark)
- Critically short telomeres signal the cell to undergo senescence or apoptosis, limiting cell division. (1 mark)
- Telomere shortening contributes to ageing and imposes a replication limit (Hayflick limit) on somatic cells. (1 mark)
Quizzes
Test 1
Test 2
Test 3
Test 4