16.14 Gene Control in Prokaryotes
1. What is Gene Expression?
- Definition:
- Gene Expression: The process by which the information encoded in a gene is used to direct the synthesis of a functional product, usually a protein.
- Stages of Gene Expression:
- Transcription: The DNA sequence of a gene is transcribed into messenger RNA (mRNA).
- Translation: The mRNA is translated by ribosomes into a specific protein.
- Illustration:
- Figure 1: Diagram showing DNA → mRNA → Protein synthesis.
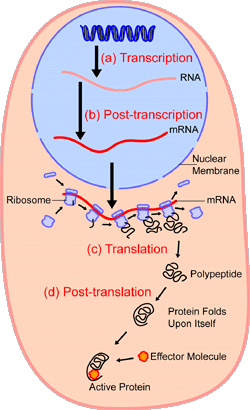
2. Selective Gene Expression
- Definition:
- Not all genes in an organism’s genome are expressed simultaneously. Selective expression ensures that only specific genes are active in a particular cell type, allowing for cellular differentiation and specialization.
- Examples:
- Melanin Production: The melanin gene is expressed in skin cells, resulting in pigment production, but remains inactive in heart muscle cells.
- Hemoglobin Synthesis: Only erythrocytes (red blood cells) express hemoglobin genes, enabling oxygen transport.
- Significance:
- Cellular Specialization: Allows diverse cell types to perform specialized functions despite having the same genetic material.
- Energy Efficiency: Conserves energy by only producing necessary proteins.
3. Gene Control in Prokaryotes
- Prokaryotic gene regulation is typically organized around operons, allowing coordinated expression of genes with related functions.
Operons in Bacteria
- Definition:
- An operon is a cluster of genes under the control of a single promoter and regulatory elements, enabling them to be transcribed together as a unit.
- Components of an Operon:
- Promoter: A DNA sequence where RNA polymerase binds to initiate transcription.
- Operator: A regulatory DNA sequence where repressor proteins can bind to block transcription.
- Structural Genes: Genes that encode proteins with related functions, transcribed together.
- Example: The lac Operon in Escherichia coli
- Function: Regulates the metabolism of lactose, allowing the bacterium to utilize lactose when glucose is not available.
- Structural Genes:
- lacZ: Encodes β-galactosidase, which breaks down lactose into glucose and galactose.
- lacY: Encodes permease, facilitating lactose entry into the cell.
- lacA: Encodes transacetylase, involved in lactose detoxification.
- Regulatory Elements:
- Promoter (P): Site for RNA polymerase binding.
- Operator (O): Binding site for the lac repressor protein.
- Regulator Gene (lacI): Located adjacent to the operon, encodes the repressor protein.
4. Structural and Regulatory Genes
- Structural Genes:
- Definition: Genes that encode proteins performing specific functions within the cell, such as enzymes, structural proteins, or transporters.
- Example: lacZ encodes β-galactosidase, an enzyme that hydrolyzes lactose.
- Regulatory Genes:
- Definition: Genes that produce proteins regulating the expression of other genes, often acting as repressors or activators.
- Example: lacI encodes the lac repressor protein, which inhibits the lac operon in the absence of lactose.
- Figure 2: Diagram differentiating structural genes and regulatory genes within an operon.
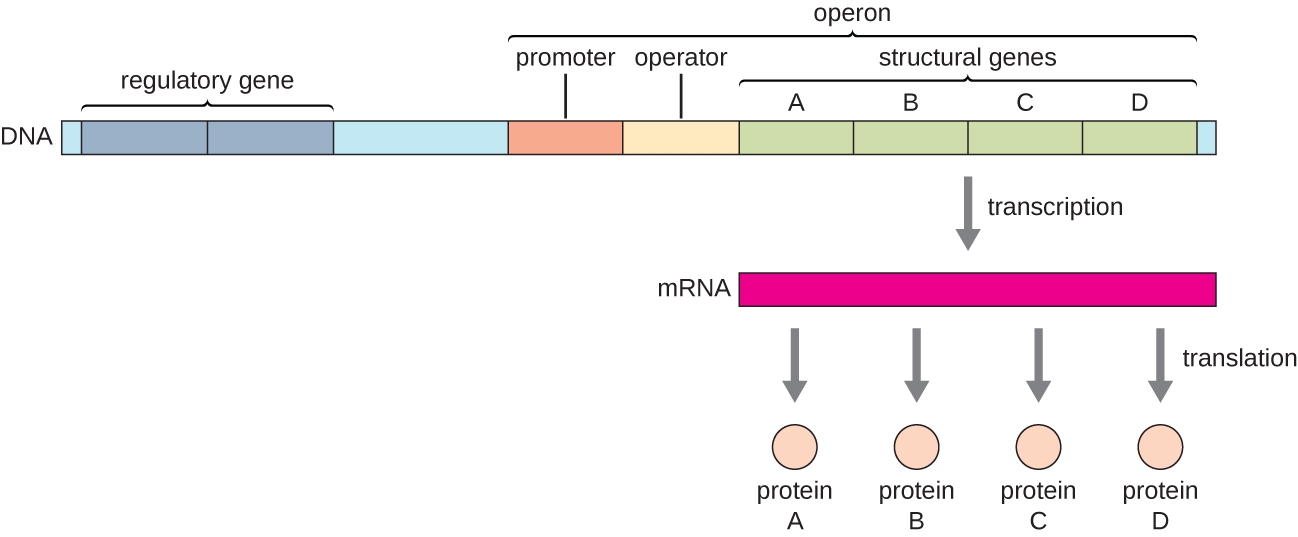
5. The lac Operon Mechanism in E. coli
- Understanding the lac operon provides insight into how bacteria regulate gene expression in response to environmental changes.
Function of the lac Operon
- Role: Controls the production of enzymes necessary for the metabolism of lactose, specifically β-galactosidase, permease, and transacetylase.
- Process: Allows E. coli to efficiently utilize lactose when glucose is scarce.
Components of the lac Operon
- lacZ: Encodes β-galactosidase.
- lacY: Encodes permease.
- lacA: Encodes transacetylase.
- Promoter (P): Site where RNA polymerase binds to initiate transcription.
- Operator (O): Binding site for the repressor protein.
Regulatory Gene: lacI
- Function: Produces the lac repressor protein, which regulates the operon by binding to the operator.
- Location: Typically located adjacent to the lac operon but is transcribed independently.
Mechanism of Action
- When No Lactose Is Present:
- Repressor Binding: The lac repressor protein binds to the operator region.
- Transcription Blocked: RNA polymerase cannot access the promoter, preventing transcription of lacZ, lacY, and lacA.
- Result: No production of β-galactosidase, permease, or transacetylase, conserving cellular resources.
- When Lactose Is Present:
- Lactose Uptake: Lactose enters the cell via permease.
- Allolactose Formation: Lactose is converted into allolactose, which acts as an inducer.
- Repressor Modification: Allolactose binds to the repressor, causing a conformational change.
- Repressor Detachment: The modified repressor can no longer bind to the operator.
- Transcription Initiated: RNA polymerase binds to the promoter, transcribing the structural genes.
- Result: Production of β-galactosidase, permease, and transacetylase, enabling lactose metabolism.
- Figure 3: Detailed mechanism of the lac operon in the presence and absence of lactose.
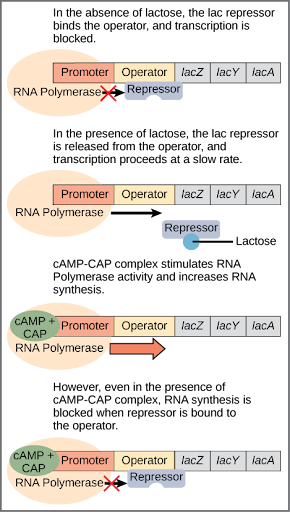
6. Types of Enzymes Based on Regulation
- Enzymes involved in metabolism can be categorized based on how their synthesis is regulated:
Inducible Enzymes
- Definition: Enzymes synthesized only in the presence of their specific substrate.
- Mechanism: The substrate acts as an inducer, activating gene expression.
- Example: β-galactosidase in the lac operon is an inducible enzyme; it is produced only when lactose is present.
Repressible Enzymes
- Definition: Enzymes that are typically produced continuously but can be inhibited when a specific effector molecule is present.
- Mechanism: The effector molecule binds to a repressor, enabling it to bind to the operator and block transcription.
- Example: Amino acid biosynthesis enzymes (e.g., tryptophan synthase) are repressible; excess tryptophan inhibits their synthesis.
- Table 1:
| Feature | Inducible Enzymes | Repressible Enzymes |
|---|
| Synthesis Trigger | Presence of substrate | Absence of end product |
| Regulatory Molecule | Inducer (e.g., lactose) | Corepressor (e.g., tryptophan) |
| Example | β-galactosidase (lac operon) | Tryptophan synthase |
| Energy Conservation | Saves energy by producing only when needed | Prevents waste by stopping production when not needed |
7. Key Terms
- β-Galactosidase: An enzyme that hydrolyzes lactose into glucose and galactose; encoded by the lacZ gene.
- Operon: A cluster of genes under the control of a single promoter and regulatory elements, allowing coordinated gene expression.
- Lac Repressor Protein: A regulatory protein encoded by the lacI gene that inhibits transcription of the lac operon in the absence of lactose.
- Inducible System: A gene regulation system activated by the presence of a specific inducer molecule.
- Repressible System: A gene regulation system inhibited by the presence of a specific corepressor molecule.
8. Example Application: Adaptive Response of the lac Operon
- The lac operon exemplifies how bacteria adapt to environmental changes to optimize resource utilization.
- Adaptive Advantage:
- Energy Efficiency: E. coli only produces lactose-metabolizing enzymes when lactose is available, conserving energy and resources.
- Rapid Response: Enables quick adaptation to fluctuations in available nutrients.
- Environmental Cue: Presence or absence of lactose in the environment triggers the operon’s regulatory mechanism.
- Figure 4: Adaptive response of the lac operon to varying lactose concentrations.
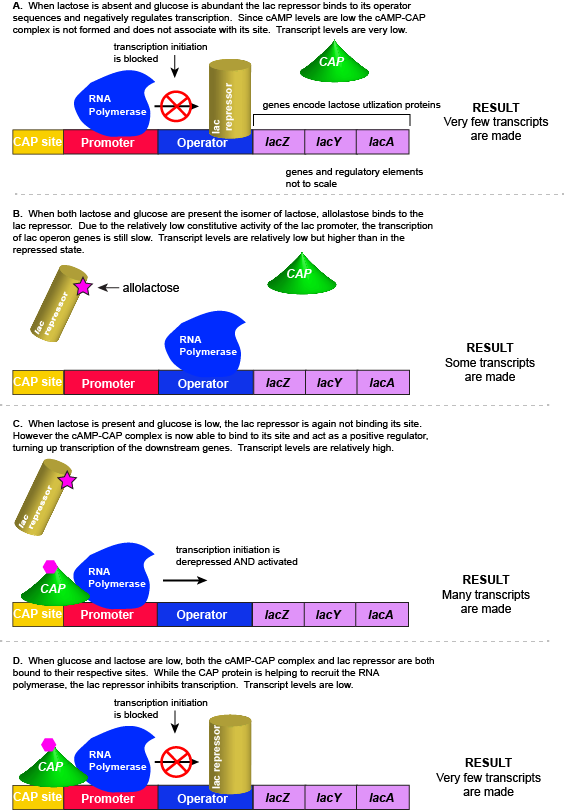
9. Comparison with Repressible Systems
Understanding different gene regulation systems highlights the versatility of cellular control mechanisms.
- Inducible Systems (e.g., lac operon):
- Activated by: Presence of substrate (e.g., lactose).
- Purpose: Enable utilization of available resources.
- Repressible Systems (e.g., trp operon):
- Activated by: Absence of end product (e.g., tryptophan).
- Purpose: Prevent overproduction and conserve resources when the end product is abundant.
- Key Differences:
| Feature | Inducible Systems | Repressible Systems |
|---|
| Trigger for Activation | Presence of substrate | Absence of end product |
| Function | Enable use of available resources | Prevent waste by stopping production |
| Example | lac operon | trp operon |